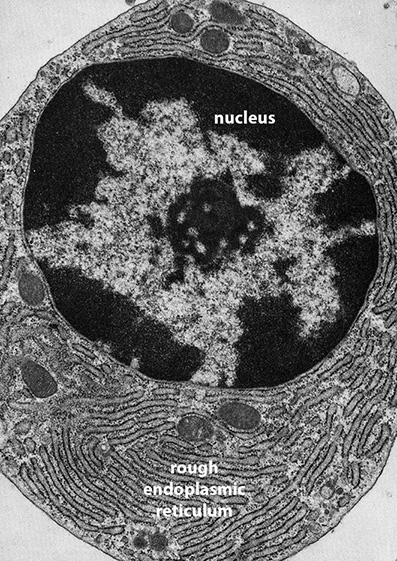
[1]
Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006 Feb 10:124(3):573-86
[PubMed PMID: 16469703]
[2]
Tannous A, Pisoni GB, Hebert DN, Molinari M. N-linked sugar-regulated protein folding and quality control in the ER. Seminars in cell & developmental biology. 2015 May:41():79-89. doi: 10.1016/j.semcdb.2014.12.001. Epub 2014 Dec 19
[PubMed PMID: 25534658]
Level 2 (mid-level) evidence
[3]
Lewy TG, Grabowski JM, Bloom ME. BiP: Master Regulator of the Unfolded Protein Response and Crucial Factor in Flavivirus Biology
. The Yale journal of biology and medicine. 2017 Jun:90(2):291-300
[PubMed PMID: 28656015]
[4]
García IA,Martinez HE,Alvarez C, Rab1b regulates COPI and COPII dynamics in mammalian cells. Cellular logistics. 2011 Jul;
[PubMed PMID: 22279615]
[5]
Bole DG, Dowin R, Doriaux M, Jamieson JD. Immunocytochemical localization of BiP to the rough endoplasmic reticulum: evidence for protein sorting by selective retention. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 1989 Dec:37(12):1817-23
[PubMed PMID: 2685110]
[6]
Sanderson CM, Crowe JS, Meyer DI. Protein retention in yeast rough endoplasmic reticulum: expression and assembly of human ribophorin I. The Journal of cell biology. 1990 Dec:111(6 Pt 2):2861-70
[PubMed PMID: 2269658]
[7]
Schwarz DS, Blower MD. The endoplasmic reticulum: structure, function and response to cellular signaling. Cellular and molecular life sciences : CMLS. 2016 Jan:73(1):79-94. doi: 10.1007/s00018-015-2052-6. Epub 2015 Oct 3
[PubMed PMID: 26433683]
[9]
Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science (New York, N.Y.). 2001 May 25:292(5521):1552-5
[PubMed PMID: 11375494]
[10]
Schaffar G, Breuer P, Boteva R, Behrends C, Tzvetkov N, Strippel N, Sakahira H, Siegers K, Hayer-Hartl M, Hartl FU. Cellular toxicity of polyglutamine expansion proteins: mechanism of transcription factor deactivation. Molecular cell. 2004 Jul 2:15(1):95-105
[PubMed PMID: 15225551]
[11]
Sugars KL, Rubinsztein DC. Transcriptional abnormalities in Huntington disease. Trends in genetics : TIG. 2003 May:19(5):233-8
[PubMed PMID: 12711212]
[12]
Hitomi J, Katayama T, Eguchi Y, Kudo T, Taniguchi M, Koyama Y, Manabe T, Yamagishi S, Bando Y, Imaizumi K, Tsujimoto Y, Tohyama M. Involvement of caspase-4 in endoplasmic reticulum stress-induced apoptosis and Abeta-induced cell death. The Journal of cell biology. 2004 May 10:165(3):347-56
[PubMed PMID: 15123740]
[13]
Onuki R, Bando Y, Suyama E, Katayama T, Kawasaki H, Baba T, Tohyama M, Taira K. An RNA-dependent protein kinase is involved in tunicamycin-induced apoptosis and Alzheimer's disease. The EMBO journal. 2004 Feb 25:23(4):959-68
[PubMed PMID: 14765129]
[14]
Kikuchi H, Almer G, Yamashita S, Guégan C, Nagai M, Xu Z, Sosunov AA, McKhann GM 2nd, Przedborski S. Spinal cord endoplasmic reticulum stress associated with a microsomal accumulation of mutant superoxide dismutase-1 in an ALS model. Proceedings of the National Academy of Sciences of the United States of America. 2006 Apr 11:103(15):6025-30
[PubMed PMID: 16595634]
[15]
Tessitore A, del P Martin M, Sano R, Ma Y, Mann L, Ingrassia A, Laywell ED, Steindler DA, Hendershot LM, d'Azzo A. GM1-ganglioside-mediated activation of the unfolded protein response causes neuronal death in a neurodegenerative gangliosidosis. Molecular cell. 2004 Sep 10:15(5):753-66
[PubMed PMID: 15350219]